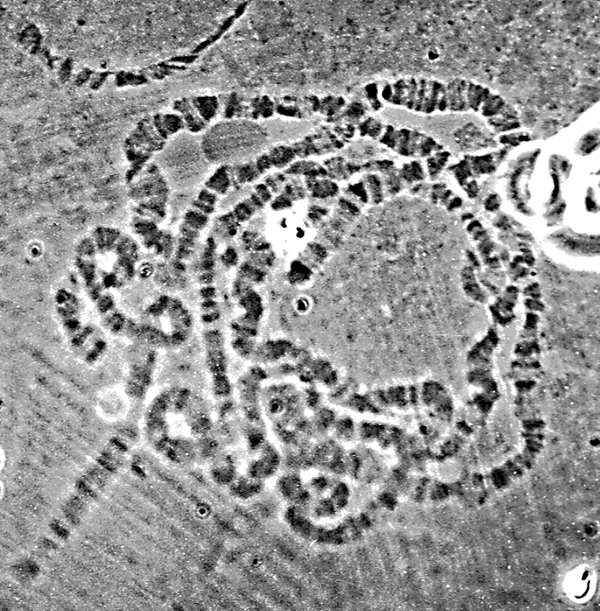
Most studies on the chromosomes of Drosophila involve the salivary gland polytene chromosomes. These extra-large chromosomes arise due to the loss of the replication block that is present in most eukaryotic cells after the cellular S phase. As a result, the salivary cells contain approximately 1000 copies of each interphase chromosome arranged side by side. These giant chromosomes are visible under the light microscope and can easily be examined in considerable detail (Figure 2).
The five Drosophila histone genes, which are the primary subject for this investigation, are located in a 5kb cluster on chromosome 2R. This cluster is tandemly repeated approximately 110 times, with few variations in either the genes or the non-expressed regions (Strausburg 1982; Matsuo and Yamazaki 1989b). The primary polymorphism lies in a 240 bp tRNA-derived addition element between the H1 and H3 genes that distinguishes the more common 'long' cluster from the 'short' cluster. The long (L) cluster sequence is stored in GenBank under accession number X14215 (Matsuo and Yamazaki 1989a). The short (S) form is 4801 bp and lacks the tRNA element. The S form, used in this project, is shown in Appendix B with annotations. Other minor conserved variants make up approximately 2% of the repeats (Liften et al 1977).
The genes in the histone cluster are sequentially arranged as shown in Figure 3. Each coding sequence is individually contained within one exon. The 16 base purine-rich target selected for this project is located in the intergenic area between H3 and H4, fifty-five residues downstream of the AvaI restriction site within this region. The target represents the terminal 16 bp of a 33 bp homopurine run. Unlike some homopurine tracts in other organisms (Giovannangeli et al 1997 for example), this tract has no known function.
The second target we have chosen to investigate in this project is the AAGAGAG heptat repeat found in Drosophila heterochromatin. Heterochromatin is a general term used to describe chromosomal regions that remain condensed during the entire cell-cycle. Functionally, they have a very low gene density much like telomeres, replicate late in S phase, and are crowded with highly repetitive DNA (reviewed in John 1988). One theory on the function of heterochromatin is that it allows chromosomal pairing and/or chromosomal recombination. However, neither of these hypotheses have been suitably proved or disproved. In Drosophila, heterochromatin makes up as much as 30% of the entire genome. Most of this DNA is spread out over eleven simple repeats that fit the pattern (RRN)m(RN)n (Lohe and Brutlag 1986).
It has been shown that the GAGA protein, the product of the Trithorax-like (Trl) gene, directly binds to AAGAGAG sequences (Farkas et al 1994), which occur in the regulatory regions of many important genes (Biggin and Tijan 1988). In studies to date, the GAGA protein functions as a transcription factor in the genes that have been analyzed. It is now postulated that rather than actively recruiting the transcription holoenzyme (RNA pol II, TBP, etc.) as do many other eukaryotic transcription factors, the GAGA protein helps to disrupt the tightly wound chromatin structure and thereby allows RNA polymerases to elongate RNA transcripts efficiently. DNase I footprinting assays on heat shock inducible genes have shown that the GAGA protein is normally found distributed throughout the regulatory regions of those genes. When heat shock is applied, the distribution of the protein extends to the entire transcriptional region, and nucleosome structure is disrupted (O'Brien et al 1995). Whether this activity is related to that of topoisomerases or helicases is unknown.
A second function of the GAGA protein was uncovered when it was shown that the protein is associated with specific heterochromatin regions throughout the cell cycle. Based upon the previous hypothesis of its activity, random generic binding by the GAGA protein should be prohibited as it is a regulatory protein.
Deletion or mutation of Trl affects "pre-cellular blastoderm nuclear division cycles" (Bhat et al 1996). In syncytial embryos of Trl- mutants, segregation of nuclei is disrupted and chromatin condensation is often disturbed by the twelfth pre-cellular division cycle. This observation, coupled to the previously characterized regulatory effects of GAGA protein, suggest that its primary activity is to modify the highly compacted and ordered chromatin structure, perhaps by disrupting the nucleosomes. We presume that binding of a third-strand to the target GAGA satellite sequences after injection into wildtype embryos should yield a similar loss of function phenotype and lead to embryonic lethality.
 |
| Figure 2. Light microscope view of stained Drosophila polytene chromosomes. The banding pattern and length of each individual chromosome can be seen. Image provided by Liz Gavis. |
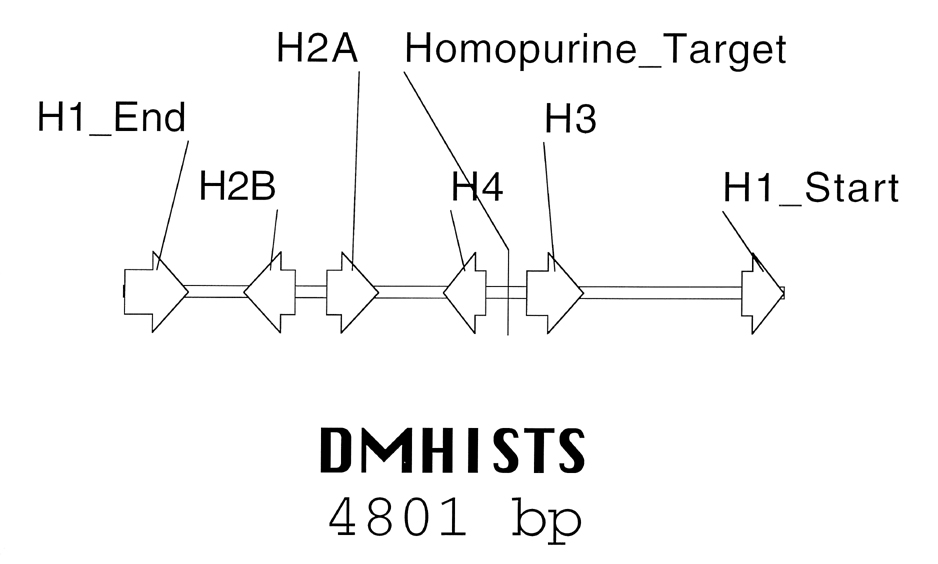 |
| Figure 3. Schematic representation of the Drosophila melanogaster histone gene cluster (S form). The generic cluster is repeated approximately 110 times in the Drosophila genome on chromosome 2. Note the location of the third-strand binding site between the H3 and H4 genes. |