The Workings Of Third-Strand Binding
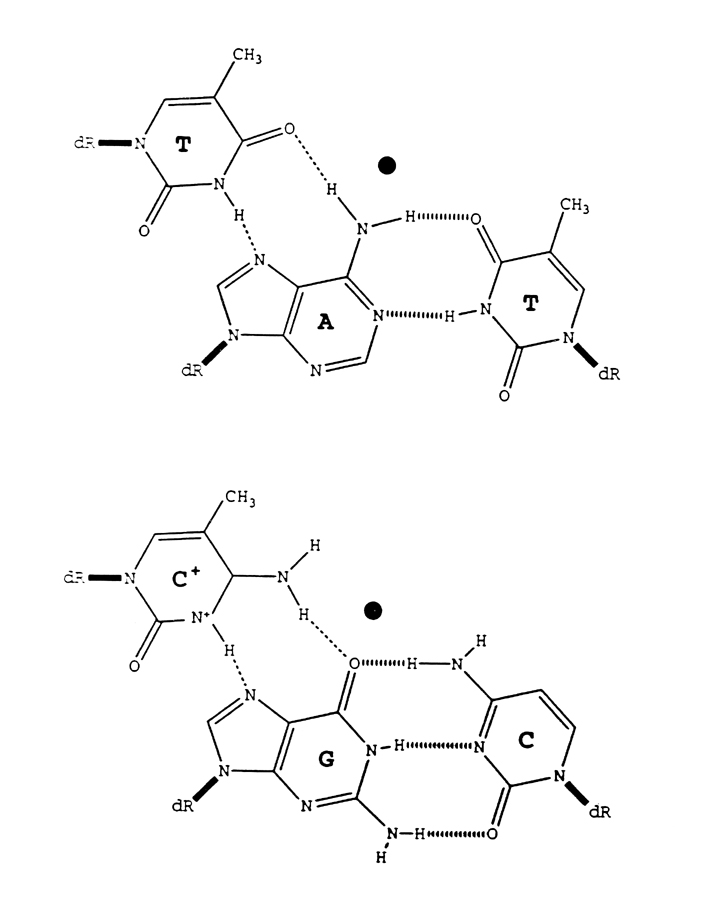
Third-strands bind to target sequences within the major groove of the DNA double helix. The bases of the incoming third-strand bind via hydrogen bonds to the exposed edge of their cognate Watson-Crick base pair (Figure 1). Structural limitations of triplex formation require a purine-rich·pyrimidine-rich duplex sequence to which the third-strand will bind. The third-strand can bind in a parallel or antiparallel orientation relative to the purine-rich target strand. The sequence specificity of third-strand binding leads to the third-strand binding code shown in Table 1.
Table 1
Table 1. Triplex helix formation is indicated by a plus (+) sign, where the polypurine strand of the duplex binds to the third-strand residues. A minus (-) sign indicates lack of binding. Third-strand cytosine residues are usually found as C+. Adapted from Letai et al (1988). |
| The Third-Strand Binding Code |
| |
|
third-strand residues |
| |
|
A |
U/T |
G |
C |
| Watson-Crick core poly-purine strand residues |
A
G |
+
- |
+
- |
-
+ |
-
+ |
By binding directly to the DNA duplex, a third-strand oligonucleotide can affect the transcription of the region to which it is bound. Cooney et al have shown that a third-strand when bound to the myc transcription factor binding site inhibits expression of the target gene (1994). Therefore, the binding interaction of this third-strand to the duplex is strong enough to compete with the binding affinity of the cell's transcription machinery. More importantly, however, the energy cost of a triplex mispair (Y:C·G or Y:T·A) has been measured to cost approximately 3-6 kcal relative to that for the canonical Y:R·Y triplets (C:G·C or T:A·T) or the complementary Watson-Crick basepairs (Griffin et al 1989). As a result of this increase in energy, approximately only one mispair can be tolerated per ten correctly paired third-strand residues (J Fresco, personal communication). This level of accuracy provides sufficient binding strength for biological applications such as those mentioned previously while avoiding non-specific binding to other sequences.
Overview of the thermodynamics of binding
The formation of a triplex DNA structure is modulated by and dependent on a variety of energetic concerns. Some of these interactions help to stabilize the new structure, while others promote disassociation of the complex.
Crystallographic data has indicated that some third-strand bound Watson-Crick duplexes take on an A-DNA configuration. However, this observation is in conflict with findings by Feigon who, using 2D NMR, has suggested a modified B-DNA structure with twelve residues per turn for the specific sequences he examined (1992). These contradictory results might be explained by a dynamic interaction at the ends of the third-strand binding region which shift the overall structure from one form to another. If true, this mechanism might have serious implications for the binding of many consecutive third-strands at closely arranged targets.
Enthalpic costs are important when a third-strand approaches the duplex for binding. The resulting fifty percent increase in the concentration of negative charges due to the insertion of a third phosphate backbone makes binding of a third-strand to a duplex weaker than that between the two strands of a duplex. Thus, melting temperatures seen for dissociation of third-strands are, depending upon third-strand length, substantially less than those observed for their target duplexes. Digression from the third-strand binding code (Fosella et al 1993) and variations in solvent conditions can also affect the observed Tm value for third-strand binding. Short oligonucleotides of less than fifteen residues usually do not bind stably and generate only minor interactions with their target. Base triplets can also provide negative influence on the affinity of binding, such as when the triplet C+:G·C is repeated multiple times. Third-strand binding is also sensitive to base inversions which interrupt homopurine·homopyrimidine target continuity and deviate from the third-strand binding code (Robertson and Crothers 1991), particularly when these base mispairs occur near the center of the structure.
As mentioned, third-strand binding strength can be changed by modifying certain solvent conditions. Higher cationic concentrations, for example, produce more effective charge shielding, alleviating the electrostatic repulsion due to the close proximity of the three phosphate backbones. Temperature can also be lowered to help stabilize triplexes. Polyamines and other positively charged ions help stabilize a triplex structure. Spermine, spermidine and benzo[e]pyridoindole help stabilize both inter- and intramolecular triplexes (Sridhara-Rao 1994).
Hydrogen bonding between bases, van der Waal interactions, and base stacking contribute to the overall energy of binding. The bases of the third-strand can also be modified to improve triplex stability. Normally, N3 of C needs to be protonated in order to form a stable Hoogsteen pair with the corresponding G in the duplex, but such protonation is energetically unfavorable at pH 7 since the intrinsic pKa of C is ~4.3. However, when C is methylated at C5, the pKa of the resulting 5-methyl-2'-deoxycytosine (m5C) is significantly higher, which enhances the binding strength to the corresponding G of the target. Therefore, lower pH is often favored in binding third-strands containing non-modified C.
Povsic and Dervan have shown that m5C increases third-strand binding affinity (1989). This effect could be due to the creation of a hydrophobic region around the triplex that generates favorable entropic effects (Xodo et al 1991). 5-propyne-2'-deoxyuridine (p5U) is another common base analog used to increase binding affinities. It is postulated that the planar prosthetic group increases base stacking interactions and, in a manner similar to m5C, increases the entropic energy of the overall structure (Froehler et al 1992).