Drosophila melanogaster was chosen as the experimental organism for this project after no suitable human sequences were found that fit the following initial guidelines:
Two possible targets in Drosophila were suggested by Dr. Paul Schedl that might fit these guidelines. The histone genes in most invertebrates are grouped together in tandem repeats. As shown below, a suitable binding target was found within the Drosophila histone cluster. This organism has the advantage that its polytene chromosomes present unique possibilities for future research.
For comparative reasons, a second Drosophila target was also chosen. The AAGAGAG septet is found as long heterochromatin repeats. The close proximity of target sites can provide additional insights into the mechanism of third-strand binding. Moreover, the role of the GAGA target in development opens the possibility for future in vivo assays.
The Drosophila histone cluster contains a 33 bp purine-rich run. A 16 bp sequence within this run was selected because it seems to provide the best set of binding interactions and contains only a single inverted base pair. The purine-rich sequence chosen, 5'-AGAGAGAAAAACAGAG-3', has one mispair (underlined), but it is devoid of repetitive guanines and long generic stretches, such as (AG)6.
The target sequence within the Drosophila GAGA satellite was chosen to encompass two full repeats (14 bp) and a fraction of a third (2 bp) for several reasons. First, the sixteen base oligomer is within the optimal size range for effective third-strand binding specificity. Furthermore, the 5 bp spacer between targets should prevent physical contact between two adjacently bound third-strands. Finally, the complementary third-strand is not cytosine-rich, and therefore favorable for binding in vivo, as it should bind tolerably well at physiological pH.
Third-strand oligodeoxynucleotides were chosen by following the third-strand binding code for parallel homopyrimidine third-strands. In the case of the histone oligomer, dp5U was used to oppose the C·G mispair. As mentioned previously, dm5C was used in place of dC. Both third strands are 16 residues long and should bind in a parallel orientation to the DNA duplex purine-rich strand, as shown in Figure 4.
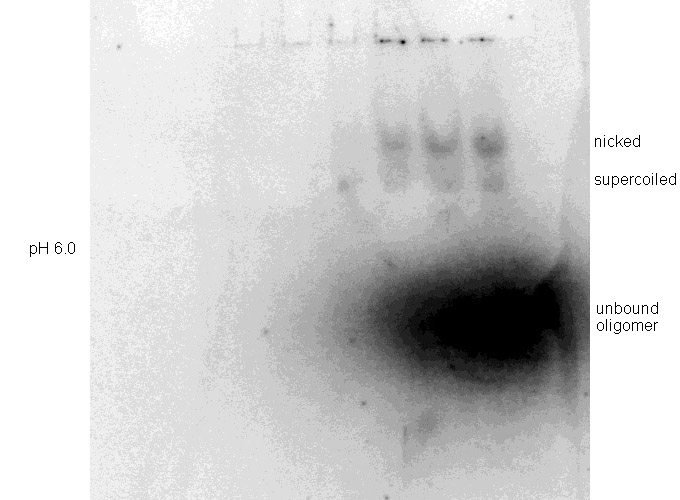
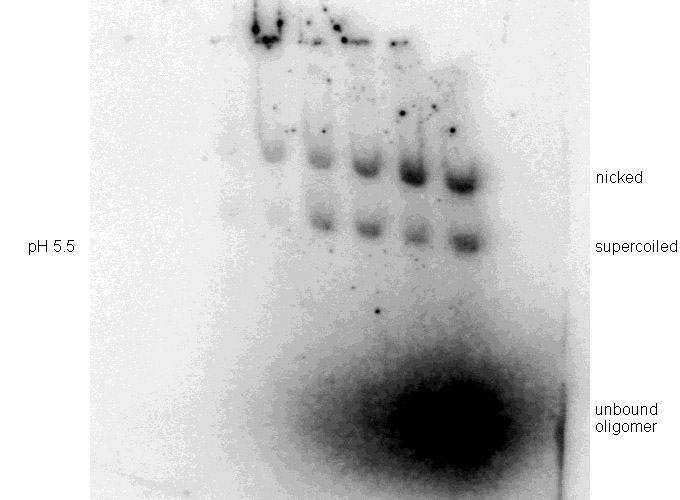
Binding experiments were performed at pH 5.5 after preliminary testing showed poor binding at pH 6.0 (Figure 5). Other reaction conditions were maintained constant as noted in the Methods.
| Third-Strand Sequence | Target DNA Sequence | Third-Strand Sequence | Target DNA Sequence | |||
5' | T T m5C+ T m5C+ T m5C+ T T m5C+ T m5C+ T m5C+ T T | 3' |
* * * * * * * * * * * * * * * * |
5' 3' | | A - T A - T G - C A - T G - C A - T G - C A - T A - T G - C A - T G - C A - T G - C A - T A - T | | 3' 5' |
5' | T m5C+ T m5C+ T m5C+ T T T T T p5U T m5C+ T m5C+ | 3' |
* * * * * * * * * * * ¤ * * * * |
5' 3' | | A - T G - C A - T G - C A - T G - C A - T A - T A - T A - T A - T C - G A - T G - C A - T G - C | | 3' 5' |
|
| GAGA | Histone | |||||
| Figure 4. Schematic representation of the third-strands binding to their target sequences. Watson-Crick base pairing is represented by dashes, Hoogsteen binding by asterisks. (Left) Drosophila GAGA satellite repeat target sequence. (Right) Drosophila histone target sequence. The base mispair is indicated by ¤. | ||||||
 |
 |
| Figure 5. Binding experiments on pMJ11 at (Top) pH 6.0 and (Bottom) pH 5.5. Binding reactions and DNA electrophoreses were done as described in Methods and the gels exposed for 35 minutes. |