Figure 6. Restriction enzyme digests of pMJ11. Approximately 2 痢 of plasmid DNA were digested with 20 units of each enzyme as indicated for 60 minutes at 37 ° C. Size markers are shown on the left.

Identification and sequence analysis of the pBR322/DMHISTS and pBR322/GAGA constructs were done using the University of Wisconsin GCG package (vers. 8.1, 9.0) on the Department of Molecular Biology (Princeton University) VAX and UNiX systems and the Vector NTI Viewer (ver. 4.0.1) created by Informax, Inc.
Plasmid pMJ10 containing the Drosophila 309 bp AAGAGAG repeat in pBR322 was kindly provided by Dr. Paul Schedl and is as described in Lohe and Brutlag (1986). pMJ10 was transformed into DH5α and transformant colonies were selected on LB/ampicillin [200 mg/L]. Two double-purified isolates of DH5α/pMJ10 were designated strains MJ31 and MJ32.
Plasmid pMJ11 consists of the E. coli vector pBR322 plus the Drosophila histone S (4.8kb form) cluster inserted at the HindIII restriction site. The original stock of pBR322/DMHISTS was kindly provided by Dr. Paul Schedl. Orientation and position of the cluster inside the vector was determined using restriction enzymes AvaI, BamHI, ClaI, HindIII, EcoRI, and XhoI (NEBiolabs) (Figure 6). Unpurified pMJ11 was transformed into DH5α, and transformants were selected on LB/ampicillin [200 mg/L]. Two double-purified isolates of DH5α/pMJ11 were designated strains MJ33 and MJ34.
Plasmid pMJ12 is a small deletion derivative of pSP64. The vector was sequentially digested with 20 units each of SmaI and HincII (NEBiolabs), yielding 2982 bp and 17 bp fragments. The 2982 bp blunt end fragment was isolated using a QiaGen Purification column. The vector was then recircularized with T4 DNA ligase (NEBiolabs) in the presence of 2.5% Ficoll for 18 hours at 16 °C. Ligated plasmid was transformed into DH5α and selected on LB/ampicillin [200 mg/L]. Rapid plasmid DNA preps were screened with BamHI for successful deletion of the 17 bp region. Two double-purified isolates of DH5α/pMJ12 were designated strains MJ35 and MJ36.
The plasmid pMJ11 was linearized with XhoI. The resulting sticky ends were digested with Mung Bean Nuclease (NEBiolabs), yielding a 9158 bp linear DNA fragment. The DNA was recircularized, transformed into DH5α, propagated and colonies purified. Rapid plasmid DNA preps were screened with AvaI for successful removal of the XhoI site. The new XhoI- plasmid was then digested with HindIII, yielding two fragments of 4361 bp and 4797 bp in length. The 4797 bp fragment is the entire XhoI- histone S cluster. The other band is linearized pBR322. The 4797 bp HindIII fragment was gel purified on low melting point agarose and isolated. It was then ligated into pMJ12 linearized at the HindIII site, designated pMJ13, and transformed into DH5α. Ampicillin resistance [200 mg/L] was used to select colonies which had taken up pMJ13. Two double-purified isolates of DH5α/pMJ13 was designated strains MJ37 and MJ38.
Strains MJ31 and MJ33 were cultured overnight in LB/ampicillin [200 mg/L]. Both cultures were treated with chloramphenicol [20 痢/mL] for two hours to amplify the plasmid. A standard 2X CsCl gradient plasmid prep protocol was used (Sanbrook et al 1989) to isolate pure pMJ10. pMJ11 was isolated using a modified Brij-Doc/QiaGen Midi Prep protocol. Resuspension was done in 1X DNA buffer (10mM Tris-Cl, 100mM NaCl, 1mM EDTA, pH 8.0). Final concentrations were estimated by ethidium bromide staining of sample titrations in an agarose gel (Figure 7).
Unpurified GAGA and histone third-strand oligodeoxynucleotides were graciously provided by Codon Pharmaceuticals (Gaithersburg, Maryland). N-1 oligomers were removed using a 20% denaturing polyacrylamide gel in 1X TBE buffer. Each complete 16 residue oligonucleotide was independently eluted into 1.0mM Tris-Cl, 0.1mM EDTA pH 7.5 solution, purified using a Waters Sep-Pak C18 cartridge, lyophilized, and resuspended into 100 無 sterile ddH20.
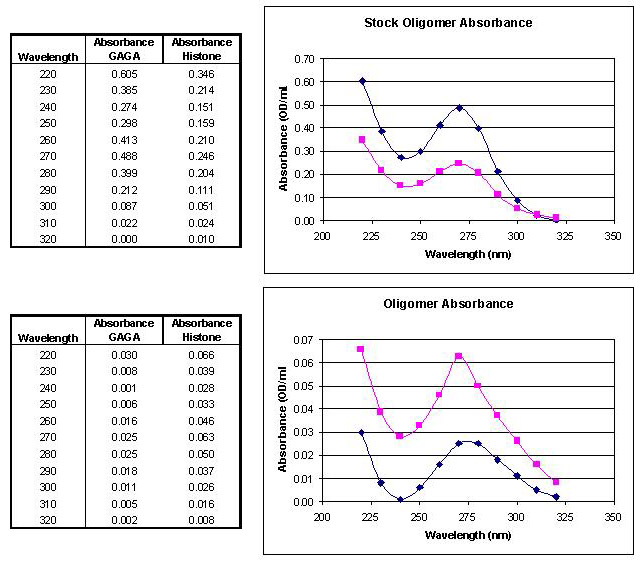
Oligonucleotide concentration was determined using a LKB Biochrom Ultraspec II spectrophotometer. Absorbance measurements were taken in sterile ddH2O (Figure 8). The concentration was calculated from the maximum absorbance using Beer's Law. Extinction coefficients used for dm5C, dT, and dp5U are 5.7, 9.6, and 10 mL/痠ol, respectively (Dawson 1983).
The purified 16 base histone oligomer was diluted 1:50 in ddH2O and kinased for 30 minutes at 37 °C (2x) with 20 units of T4 polynucleotide kinase (NEBiolabs) and 6000 Ci/mmol γ-32P-ATP (Amersham). 9.37 picomoles of oligomer were usually end-labeled per reaction. The labeled oligomer was purified as above, and resuspended in sterile ddH2O. Radioactive counts (cpm) were measured in the eluates before and after column purification to estimate recovery rates. 70%-85% recovery was consistently achieved.
0.066 picomoles (4×1010 molecules, final concentration = 2.66 nanomolar) of pMJ10 and pMJ11 (0.4 and 0.2 痢, respectively) were separately mixed in filtered triplex cocktail (10mM Bis-TRIS, 1mM EDTA, 10mM MgCl2, 1mM DTT, 50mM KCl, 1然 spermine, 2% PEG) with the appropriate 32P-end-labeled oligonucleotide at various oligomer:plasmid ratios (1:1, 2.5:1, 5:1, 10:1, 20:1, 40:1, 50:1) and incubated for at least 20 hours at 23 °C in a final volume of 25 無. Reaction mixtures were then run on a vertical 0.8% Seakem Gold filtered agarose gel in filtered, recirculated 1X Bis-Tris buffer (0.04M Bis-Tris, 0.04 Acetate, 0.05M MgOAc, pH 5.5) for 24 hours at 20V, 20-35mA, 23 °C. Gels were visualized via ethidium bromide staining, soaked for 20 minutes in 7% trichloroacetic acid, dried at 65 °C for 90 minutes, exposed on a Molecular Dynamics PhosphoImager for 20-40 minutes and quantitated on the Molecular Dynamics ImageQuant program (vers. 3.3 and 4.1).
Figure 6. Restriction enzyme digests of pMJ11. Approximately 2 痢 of plasmid DNA were digested with 20 units of each enzyme as indicated for 60 minutes at 37 ° C. Size markers are shown on the left. |
 |
Figure 7. Sequential volumes of purified pMJ11. 1, 2, 4, 6, and 10 無 of pMJ11 were added to each well as shown on top. Plasmid concentration was determined to be 0.1 痢/無. |
 |
Figure 8. UV absorption spectrum of purified oligomers. (Top) Base stock oligomers. [GAGA] = 3.748 pmol/無; [Histone] = 1.829 pmol/無. (Bottom) Experimental stock oligomers. [GAGA] = 0.192 pmol/無; [Histone] = 0.468 pmol/無. ◆ GAGA oligomer absorbance curve; ■ histone oligomer absorbance curve. |
 |